Solomon K. Langat1, Genay Pilarowski2, Samson Limbaso1, Paul Oluniyi2, Daniele Jones2, Albert Nyunja1, Edith Koskei1, Samuel Owaka1, Hellen Koka1, Francis Mulwa1, James Mutisya1, Victor Ofula1, Edith Chepkorir1, Joel Lutomiah1, Cristina M. Tato2, Samoel Khamadi1.
Affiliations
- Centre for Virus Research, Kenya Medical Research Institute (KEMRI), Nairobi, Kenya
- Chan Zuckerberg Biohub, San Francisco, CA, United States of America
Introduction
Hepatitis A is a vaccine-preventable liver disease caused by the hepatitis A virus (HAV). It is the sole member species of the genus Hepatovirus, family picornaviridae. HAV is an ancient human pathogen that was previously recovered only from primates. It is the causative agent of water and food-borne hepatitis (1). The virus is widely distributed across the world, and it has been implicated in several epidemics mainly in low- and middle-income countries (2,3). It is estimated that approximately 90% of children in developing countries are infected by the virus before the age of 10 years (4). In Kenya, a cross-sectional study on 300 children found an elevated exposure of HAV infection by the age of 14: approximately 77%, among children from low socio-economic backgrounds (5). Despite the public health threat posed by HAV infection, there is paucity of knowledge on the incidences and distribution of HAV infection across the world and especially in developing countries. This lack of evidence has in turn contributed to a lack of implementation of measures to curb the diseases such as the institution of vaccination programs. In this study, we utilized metagenomic next-generation sequencing (mNGS) to characterize the pathogen landscape among febrile patients who presented to different health facilities in Kenya, during a Yellow fever virus (YFV) surveillance program. Here, we report the identification and full genome characterization of HAV detected during this period.
Methods
Samples: Serum samples used in the study were YFV-suspected samples collected as part of the national YFV surveillance program, under the Division of Disease Surveillance and Response System in the Kenyan Ministry of Health. The samples were collected from persons who presented to local health facilities with febrile illnesses, and symptoms consistent with YFV infection. The collected samples were transported in cold storage to the laboratory at Kenya Medical Research Institute (KEMRI) where they were processed for serological and PCR testing.
Metagenomic next-generation sequencing (mNGS): RNA was extracted directly from serum using QIAamp Viral RNA Mini Kit, following the manufacturer’s recommended protocol without the addition of carrier RNA. The extracted RNA, along with extraction controls and water controls, was used as input for mNGS library preparation and sequencing at Chan Zuckerberg Biohub San Francisco. Libraries were prepared using the NEBNext Ultra II RNA library prep kit for Illumina (NEB, UK) with minor modifications, and the 96 prepared libraries were sequenced on a NextSeq550 with a High Output Kit in a 2x146bp paired-end configuration.
Sequence analysis: Raw sequence reads (fastq files) were uploaded to the CZID platform (https://czid.org/) for host and quality filtering, de novo assembly as well as taxonomic classification of the reads and contigs by querying against the NCBI nucleotide (NT) and non-redundant protein (NR) databases (6) using the Metagenomic module v8.3.0. Ten (10) water controls used during sequencing were selected in CZ ID to create a mass-normalized background model for use in filtering background contamination in the respective samples using a background Z-score ≥1. Additionally, only the hits that had ≥10 nucleotide reads/million and ≥5 protein reads/million mapping to specific taxa, as well as those that form an average nucleotide alignment of ≥50 base pairs were considered true hits. Contigs belonging to HAV in the different samples were downloaded from CZ ID, then compared to publicly available sequences using BLASTn, with default parameters. To generate full genomes for subsequent analysis, HAV-positive samples were run through the CZ ID Consensus genome pipeline with MH577313.1 as a reference, as it was one of the top hits across all genomes based on BLAST analysis.
Phylogenetic Analysis: Maximum likelihood phylogeny was constructed based on the complete and near-complete genome sequences (≥90% coverage) from this study and on sequences obtained from the Genbank database through NCBI Virus portal. The reference sequences were randomly selected using the random selection tool in NCBI Virus portal, and the selected sequences were stratified per country such that a maximum of 10 representative HAV sequences from each country were downloaded. These sequences included all 6 genotypes of HAV virus. The sequences were combined with those obtained from the study and the combined set were aligned using MAFFT (7) and manually edited in Aliview. The dataset was then used to infer maximum likelihood phylogeny using IQtree (8), and the generated tree was visualised in Figtree v1.4.4.
Data Availability: Host-filtered reads for the samples analyzed in the study have been submitted to sequence read archive under the Bioproject accession numbers [numbers]. The genome sequences of HAV obtained from the study are available in Genbank under accession numbers: [numbers].
Results and Discussion
A total of 37 yellow-fever suspect samples were processed for sequencing. Out of these, seven (7) samples (18.9%) were positive by mNGS for Hepatitis A virus. All seven HAV-positive samples had tested negative for yellow fever by serology and PCR. Out of the 7 individuals that tested positive for HAV, 5 were male and two were female (Table 1). These individuals were of varying age-ranges from 2 to 34 years of age.
Near-complete genomes of HAV (>90%) were recovered from five (5) of the isolates (Table 1). Partial HAV genomes, 60.07% and 30.92%, were recovered from two of the samples.
BLAST analysis based on the polyprotein coding region showed high similarities of the sequenced isolates to those available in Genbank, with three isolates showing ~97.7% nucleotide identity scores to HAV strain that was detected among patients in 2019 in New York, US (Table 1). The remaining four isolates had a high nucleotide percentage identity score to HAV strains detected among patients in the neighbouring country of Uganda (Table 1).
Table 1: Details of the sequences of HAV detected in the study.
|
Sample Information Summary |
Sequencing Information Summary |
|||||||
|
Sample_ID |
Age |
Gender |
County |
Collection Date |
Length |
Coverage (%) |
Closest Hit (Country) |
Percent Identity |
|
RR176035 |
7 |
M |
Isiolo |
29.07.2022 |
7395 |
98.89 |
MH577313 (USA) |
97.84 |
|
RR176024 |
34 |
M |
Isiolo |
10.03.2022 |
7390 |
98.82 |
OQ077985 (Uganda) |
97.02 |
|
RR176036 |
3.5 |
M |
Isiolo |
29.07.2022 |
7324 |
97.94 |
ON524437 (USA) |
97.97 |
|
RR176017 |
13 |
M |
Isiolo |
06.03.2022 |
7270 |
97.22 |
ON524426 (USA) |
97.72 |
|
RR176030 |
7 |
F |
Wajir |
29.07.2022 |
6783 |
90.71 |
OQ077985 (Uganda) |
97.1 |
|
RR176027 |
29 |
F |
Nakuru |
29.07.2022 |
4492 |
60.07 |
MH685714 (Uganda) |
98.03 |
|
RR176033 |
2 |
M |
Isiolo |
26.07.2022 |
2312 |
30.92 |
OQ077984 (Uganda) |
95.7 |
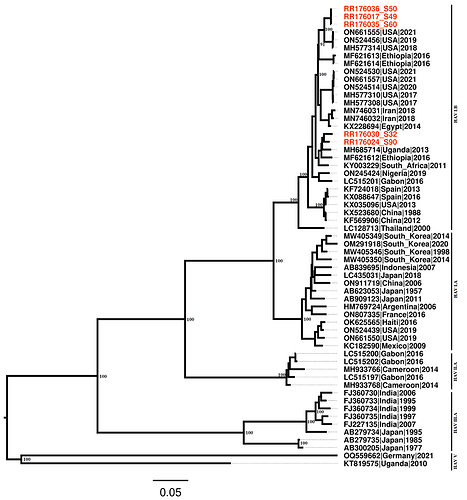
Phylogenetic analysis showed two distinct clusters of the isolates from Kenya, confirming the BLAST alignment that showed four isolates closely related to strains from Uganda and the remaining three isolates closely related to strains detected in 2019 in New York, US (Fig. 1). Further, the phylogenetic analysis placed all seven isolates under HAV genotype I.B, which is one of the genotypes commonly associated with human illnesses. The isolates in this genotype included those from around the region such as Uganda, Ethiopia, Gabon as well as Nigeria suggesting a possible cross-border transmission of this pathogen. In addition, HAV isolates from outside the region including those from the USA, Iran, Spain as well as China also clustered around the isolates detected in the study.
Fig. 1: Maximum likelihood phylogeny of the newly sequenced isolates (in red) in the context of other global strains.
Conclusion
The findings in this study reveal the circulation of HAV among febrile patients in Kenya. The study demonstrates the successful use of unbiased mNGS to detect HAV in undiagnosed febrile cases during the Yellow-fever surveillance programme. This, therefore, shows the utility of mNGS in understanding the causative pathogens of undiagnosed febrile cases to better inform the diagnostics, case management as well as the disease surveillance programs. The findings in this study highlights the need for more concerted efforts in the area of pathogen detection of HAV in Kenya in order to determine the prevalence across the different countries, and also to initiate case management among the patients. Most importantly, a revision of the National vaccination program guidelines should be considered to ensure Hepatitis A vaccination is included, especially in high incidence areas.
Acknowledgments:
We thank the Kenyan ministry of health for their support. We are also grateful to the teams from the Arbovirus and Viral Hemorrhagic Fever laboratory at KEMRI, and the Rapid Response and Genomics teams at the Chan Zuckerberg Biohub SF and Chan Zuckerberg Initiative.
This work was supported by funding from the Bill & Melinda Gates Foundation in collaboration with the Chan Zuckerberg Initiative (CZI) and Chan Zuckerberg Biohub (CZB) (INV-050635).
Data Availability:
Sequence alignments used in the study, including the newly generated sequences, and the trees generated are available in github under the following link: https://github.com/sklangat/HAV-Metagenomics
Reference
- Vaughan, G., Rossi, L. M. G., Forbi, J. C., de Paula, V. S., Purdy, M. A., Xia, G., & Khudyakov, Y. E. (2014). Hepatitis A virus: host interactions, molecular epidemiology and evolution. Infection, Genetics and Evolution, 21, 227-243.
- Jacobsen, K. H., & Wiersma, S. T. (2010). Hepatitis A virus seroprevalence by age and world region, 1990 and 2005. Vaccine, 28(41), 6653-6657.
- World Health Organization. (2000). Hepatitis A vaccines: WHO position paper. Weekly Epidemiological Record= Relevé épidémiologique hebdomadaire, 75(05), 38-44.
- World Health Organization. (2023, July 20). Hepatitis A. World Health Organization. https://www.who.int/news-room/fact-sheets/detail/hepatitis-a
- Wasunna, A., Murila, F., Obimbo, M. M., Rama, M. J., & Musembi, H. (2016). Hepatitis A antibody seroprevalence in a selected Kenyan pediatric population. Open Journal of Pediatrics, 6(4), 316.
- Kalantar, K. L., Carvalho, T., de Bourcy, C. F., Dimitrov, B., Dingle, G., Egger, R., ... & DeRisi, J. L. (2020). IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience, 9(10), giaa111.
- Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution, 30(4), 772-780.
- Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., & Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution, 32(1), 268-274.