Phylodynamic Analyses based on 11 genomes from the Italian outbreak
Insights on the R0, the number of cases through time, and the time of epidemic origin
Jérémie Sciré1,2, Timothy G. Vaughan1,2, Sarah A. Nadeau1,2, and Tanja Stadler1,2,*
1Department of Biosystems Science and Engineering, ETH Zürich, Basel, Switzerland
2Swiss Institute of Bioinformatics (SIB), Switzerland
*Correspondence to be addressed to: [email protected]
March 6th 2020
The Italian COVID-19 epidemic was seeded at least twice (NextStrain Situation Report 2020-03-04). One introduction of COVID-19 into Italy led to an outbreak with 11 sampled sequences (NextStrain). We highlight that only 1 patient from this Italian outbreak was sampled in Italy, all other patients were sampled abroad. In this report, we outline phylodynamic analyses based on full-length SARS-CoV-2 genomes downloaded from GISAID. Acknowledgements for and details of the genome sequences are given in the table at the end of this post.
In particular, we performed Bayesian phylodynamic analyses in BEAST2 [1] in order to estimate the R0 for the Italian epidemic, the total number of cases through time, as well as the time of epidemic origin.
Method
We downloaded 10 sequences from COVID-19 patients with known travel history to Italy and 1 sequence collected in Italy from GISAID. These sequences were chosen because they cluster together in the NextStrain tree and thus we hypothesise that they belong to the same Italian outbreak. We aligned the sequences using the NextStrain pipeline. In accordance with the pipeline, we masked the first 130 sites, last 50 sites, and sites 18529, 28881, 28882, and 28883 from the alignment. We refer to the resulting alignment as the “Italian-outbreak” alignment.
We apply the birth-death framework [2] to this alignment in order to quantify R0, the time of origin of the considered Italian epidemic, as well as the total number of cases until the time of the last sequence on Mar. 3. We also apply EpiInf [3] to the same data. This tool assumes the same birth-death model, and additionally provides estimates for the number of COVID-19 cases through time for the considered Italian outbreak. The prior settings for both analyses are shown in Table 1. Since 10 out of 11 of the sequences were sampled outside of Italy, the sampling procedure is “migration from Italy to abroad followed by sequencing”. We argue that such a sampling procedure ensures a “uniform at random” sampling from the Italian epidemic. Such a “uniform at random” sampling procedure is assumed by the birth-death models.
| Model | Parameter | Prior distribution |
|---|---|---|
| Strict clock | clock rate | fixed to 5 x 10-4; 10-3 |
| HKY | kappa | lognormal(1.0, 1.25) |
| Gamma shape | Exponential(0.5) | |
| Birth-death | R0 | lognormal(0.8, 0.5) |
| removal rate δ | fixed to 365/8; 365/10; 365/12 per year | |
| sampling proportion s | Unif(0, 0.05) | |
| time of origin tOrigin | lognormal(-2, 0.8) |
Table 1: Models and prior distributions used for the BEAST2 analyses. The birth-death model is described in [2], all other models are part of the core of BEAST2 [1]. We highlight that 1/δ is the time of an individual being infected, i.e. the sum of the exposed period and the infectious period. Sampling is assumed to coincide with removal from the infectious pool. For the EpiInf analyses, we put an upper bound of Jan. 1 for the time of origin.
Results
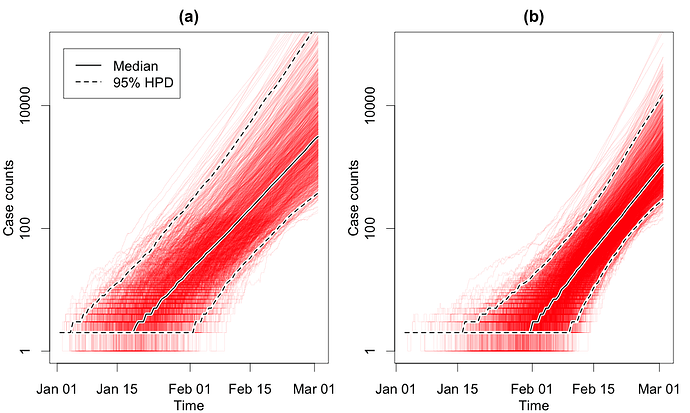
In Fig. 1, we show the number of cases through time obtained through the EpiInf analyses. In Fig. 2, we show R0 and the total number of cases until the time of the last sequence on Mar. 3.
Figure 1: Number of COVID-19 cases through time in the considered Italian outbreak. Panel (a) shows results based on a clock rate of 5 x 10-4 and panel (b) shows results based on a clock rate of 10-3. Here, we fixed δ = 365/10. The estimated number of cases on Mar. 3 was 3’136 (95% HPD 375 - 222’000) for (a) and 570 (95% HPD 1’097 - 15,831) for (b) .
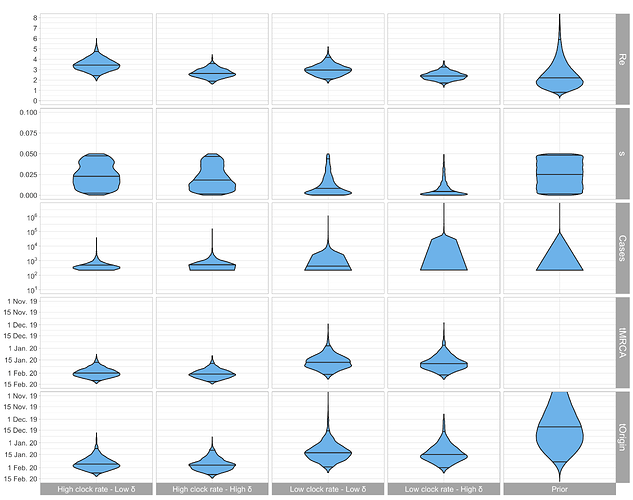
Figure 2: Posterior distribution of R0, sampling proportion s, total number of cases until Mar. 3 (= # sequences / s), tMRCA, and tOrigin estimated based on the Italian-outbreak alignment. Low δ corresponds to 365/12 and high δ to 365/8 (i.e. the sum of the exposed period and the infectious period is assumed to be 12 and 8 days, respectively).
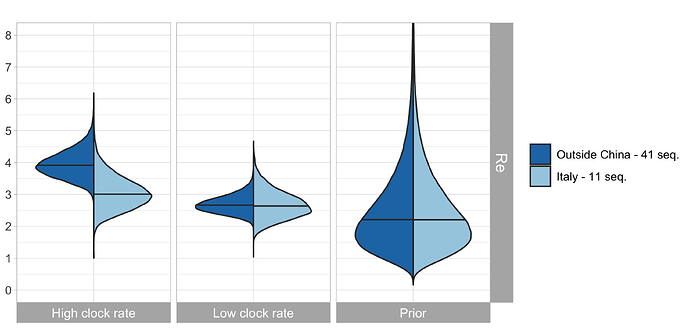
In our previous work, we quantified R0 for the initial Chinese epidemic outbreak using equivalent procedures to those described here. In particular, the R0 was estimated based on sequences collected outside of China (called the outside-China alignment containing 41 sequences), with the corresponding patients having had contacts to China. Thus, again, the sampling procedure was “migration from China to abroad followed by sequencing”. We compare the obtained R0 for China and Italy, see Fig. 3.
Figure 3: Comparison of the R0 estimate for China (based on the outside-China alignment in our previous work) and the R0 estimate for Italy (based on our Italy-outbreak alignment). This plot assumes δ = 365/10.
Summary
Based on results in our previous post, we are more confident in a clock rate of 5 x 10-4 compared to a faster clock. For this clock rate, we estimate the start of the considered Italian outbreak between mid-December and end of January (Fig. 2, bottom row). There is not yet much information on the total number of cases (Fig. 2, third row). The median estimated number of cases on Mar. 3 is around 3’136, albeit with a large credible interval. Despite the few sequences we quantify the R0 in Italy to be very similar to the R0 in China (Fig. 3, middle).
Acknowledgments
We again gratefully acknowledge the Authors and the Originating and Submitting Laboratories for their sequence and metadata shared through GISAID, on which this research is based. Below is a table containing the Italian cluster sequences used for the analysis. Information for the outside-China sequences is given in our previous post.
Click to show sequence information and acknowledgements table
| Accession ID | Virus name | Location | Collection date | Originating lab | Submitting lab | Authors |
|---|---|---|---|---|---|---|
| EPI_ISL_412912 | BetaCoV/Germany/Baden-Wuerttemberg-1/2020 | Europe / Germany / Baden-Wuerttemberg | 2020-02-25 | State Health Office Baden-Wuerttemberg | Charite_ Universitätsmedizin Berlin, Institute of Virologyh | Victor M Corman, Julia Schneider, Barbara Mu_hlemann, Talitha Veith, Jörn Beheim-Schwarzbach, Terry Jones, Rainer Oehme, Silke Fischer, Christian Drosten |
| EPI_ISL_412964 | BetaCoV/Brazil/SPBR-01/2020 | South America / Brazil / Sao Paulo / Sao Paulo | 2020-02-25 | Hospital Israelita Albert Einstein | Instituto Adolfo Lutz Interdisciplinary Procedures Center Strategic Laboratory | Jaqueline Goes de Jesus, Claudio Tavares Sacchi, Daniela Bernardes Borges da Silva, Ingra Morales Claro, Flávia Cristina da Silva Sales, Claudia Regina Gonçalves, Joshua Quick, Maria do Carmo, Sampaio Tavares Timenetsky, Nicholas James Loman, Andrew Rambaut, Ester Cerdeira Sabino, Nuno Rodrigues Faria |
| EPI_ISL_412971 | BetaCoV/Finland/FIN-25/2020 | Europe / Finland / Helsinki | 2020-02-25 | HUS Diagnostiikkakeskus, Hallinto | Department of Virology Faculty of Medicine, Medicum University of Helsinki | Teemu Smura, Suvi Kuivanen, Hannimari Kallio-Kokko, Olli Vapalahti |
| EPI_ISL_412973 | BetaCoV/Italy/CDG1/2020 | Europe / Italy / Lombardy | 2020-02-20 | Department of Infectious Diseases, Istituto Superiore di Sanità, Roma , Italy | Virology Laboratory, Scientific Department, Army Medical Center | Paola Stefanelli, Stefano Fiore, Antonella Marchi, Eleonora Benedetti, Concetta Fabiani, Giovanni Faggioni, Antonella Fortunato, Riccardo De Santis, Silvia Fillo, Anna Anselmo, Andrea Ciammaruconi, Stefano Palomba, Florigio Lista |
| EPI_ISL_412972 | BetaCoV/Mexico/CDMX/InDRE_01/2020 | North America / Mexico / Mexico City | 2020-02-27 | Instituto Nacional de Enfermedades Respiratorias | Instituto de Diagnostico y Referencia Epidemiologicos (INDRE) | Ramirez-Gonzalez Ernesto, Garces-Ayala Fabiola, Araiza-Rodriguez Adnan, Mendieta-Condado Edgar, Rodriguez-Maldonado Abril, Wong-Arambula Claudia, Vazquez-Perez Joel, Martinez Arturo, Boukadida Celia, Munoz-Medina Esteban, Sanchez Alejandro, Isa Pavel, Taboada Blanca, Lopez Susana, Arias Carlos, Barrera-Badillo Gisela, Hernandez-Rivas Lucia, Lopez-Martinez Irma |
| EPI_ISL_413020 | BetaCoV/Switzerland/1000477377/2020 | Europe / Switzerland / Zurich | 2020-02-27 | Department of Internal Medicine, Triemli Hospital | Institute of Medical Virology, University of Zurich | Stefan Schmutz, Maryam Zaheri, Verena Kufner, Patrick Redli, Fiona Steiner, Jon Huder, Riccarda Capaul, Andrea Zbinden, Jürg Böni, Michael Huber, Gerhard Eich, Alexandra Trkola |
| EPI_ISL_413021 | BetaCoV/Switzerland/1000477757/2020 | Europe / Switzerland / Zurich | 2020-02-29 | Klinik Hirslanden Zurich | Institute of Medical Virology, University of Zurich | Stefan Schmutz, Maryam Zaheri, Verena Kufner, Gabriela Ziltener, Patrick Redli, Fiona Steiner, Jon Huder, Riccarda Capaul, Andrea Zbinden, Jürg Böni, Michael Huber, Christian Ruef, Alexandra Trkola |
| EPI_ISL_413022 | BetaCoV/Switzerland/1000477796/2020 | Europe / Switzerland / Zurich | 2020-02-29 | Division of Infectious Diseases, University Hospital Zurich | Institute of Medical Virology, University of Zurich | Stefan Schmutz, Maryam Zaheri, Verena Kufner, Gabriela Ziltener, Patrick Redli, Fiona Steiner, Jon Huder, Riccarda Capaul, Andrea Zbinden, Jürg Böni, Michael Huber, Roberto Speck, Alexandra Trkola |
| EPI_ISL_413023 | BetaCoV/Switzerland/1000477797/2020 | Europe / Switzerland / Zurich | 2020-02-29 | Division of Infectious Diseases, University Hospital Zurich | Institute of Medical Virology, University of Zurich | Stefan Schmutz, Maryam Zaheri, Verena Kufner, Gabriela Ziltener, Patrick Redli, Fiona Steiner, Jon Huder, Riccarda Capaul, Andrea Zbinden, Jürg Böni, Michael Huber, Roberto Speck, Alexandra Trkola |
| EPI_ISL_413024 | BetaCoV/Switzerland/1000477806/2020 | Europe / Switzerland / Zurich | 2020-02-29 | Division of Infectious Diseases, University Hospital Zurich | Institute of Medical Virology, University of Zurich | Stefan Schmutz, Maryam Zaheri, Verena Kufner, Gabriela Ziltener, Patrick Redli, Fiona Steiner, Jon Huder, Riccarda Capaul, Andrea Zbinden, Jürg Böni, Michael Huber, Roberto Speck, Alexandra Trkola |
| EPI_ISL_413221 | BetaCoV/Scotland/CVR01/2020 | Europe / Scotland | 2020-03-02 | West of Scotland Specialist Virology Centre, NHSGGC | MRC-University of Glasgow Centre for Virus Research | Emma Thomson, Antonia Ho; James Shephard, Shirin Ashraf; Kathy Smollett, Daniel Mair, Stephen Carmichael, Ana da Silva Filipe; Richard Orton, Josh Singer, David L Robertson; Andrew Rambaut; Alasdair MacLean, Rory Gunson. |
References
- Remco Bouckaert, Timothy G. Vaughan, Joelle Barido-Sottani, Sebastián Duchêne, Mathieu Fourment, Alexandra Gavryushkina, Joseph Heled, Graham Jones, Denise Kühnert, Nicola De Maio, et al. Beast 2.5: An advanced software platform for bayesian evolutionary analysis. PLoS computational biology, 15(4):e1006650, 2019.
- Tanja Stadler, Denise Kühnert, Sebastian Bonhoeffer, and Alexei J. Drummond. Birth–death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proceedings of the National Academy of Sciences, 110(1):228–233, 2013.
- Timothy G. Vaughan, Gabriel E. Leventhal, David A. Rasmussen, Alexei J. Drummond, David Welch, and Tanja Stadler. Estimating Epidemic Incidence and Prevalence from Genomic Data. Molecular Biology and Evolution, 36(8):1804–1816