On August 15th, 2023, the Chad Ministry of Public Health and Prevention has declared an outbreak of dengue virus (DENV) in the health district of Abéché, Ouaddaï province in eastern Chad [1]. As reported on 19th December 2023, there have been 2037 suspected cases, including 100 DENV-confirmed cases across four provinces (N’Djamena (n=49), Ouaddaï (n=33), Wadi Fira (n=12), and Sila (n=6)).
As part of the Africa CDC - Africa Pathogen Genomics Initiative (Africa PGI) regional sequencing program for priority pathogens and use-cases, a Chadian research team from the National Biosafety and Epidemics Laboratory (LaBiEp) travelled on 27th November 2023, with DENV-positive samples, to the regional sequencing hub at National Institute of Biomedical Research (INRB), Kinshasa (Democratic Republic of the Congo, DRC). The whole-genome sequencing (WGS) was conducted using Oxford Nanopore Technologies (ONT) platforms at INRB. An amplicon-based sequencing approach (1500 bp amplicon) was performed using a newly designed and specific primers for DENV [2]. Out of 20 RNA-positive samples, 17 were sequenced using ONT kits and devices. We successfully obtained 4 full-length and 13 partial genomes of DENV strains circulation in Chad in 2023.
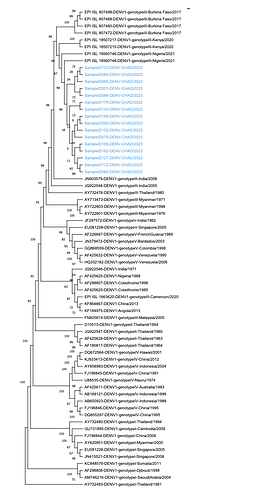
Our Maximum-Likelihood phylogenetic analysis revealed that the 2023 sequenced cases of DENV in Chad belong to DENV-1 genotype III (Figure 1).
Figure 1 : Maximum-likelihood phylogenetic tree of dengue virus, including 16 DENV-1 sequences from this study, and 53 additional DENV-1 sequences with large geographic distribution from NCBI GenBank and GISAID. The tree was inferred by using the maximum-likelihood method and general time-reversible model, with a bootstrap value of 500. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.2828)). Sequences used in this study are named as follows: ‘GenBank or GISAID accession number’-‘DENV type’-‘genotype’-‘country/year of isolation’. Sequences from the 2023 dengue outbreak in Chad are highlighted in blue and identified as follows: ‘Sample ID number’-‘DENV’-‘CHAD’. Evolutionary analyses were conducted in MEGA11 [3].
DENV type-1 genotype III sequences from this study cluster together into one cluster, with 4 sublineages, and the cluster is closely related to dengue viruses identified in Nigeria in 2021, in Kenya in 2020, and in Burkina Faso in 2017 with 99% bootstrap node support. To our knowledge, this is the first report about the serotype of DENV circulating in Chad in 2023.
Partners and collaborators
Djamal Hachim-Abdoulaye 1,#
Adrienne Amuri-Aziza 2,#
Gradi Luakanda-Ndelemo 2,#
Akil Bandali-Prince 2
Houlkeurbe Ngam-Daita 1
Anouar Mahamat-Moustaph 1
Emmanuel Lokilo-Lofiko 2
Dorcas Waruguru-Wanjohi 3
Eddy Kinganda-Lusamaki 2,4
Jean-Claude Makangara-Cigolo 2,4
Jean-Jacques Muyembe-Tamfum 2,4
Gerald Mboowa 3
Placide Mbala-Kingebeni 2,4
Oumar Ouchar-Mahamat 1
Tony Wawina-Bokalanga 2,4
1. Laboratoire National de Biosécurité et d’Épidémies (LaBiEp), N’djamena, Chad
2. Department of Epidemiology and Global Health, National Institute of Biomedical Research (INRB), Kinshasa, Democratic Republic of the Congo
3. Africa CDC, Addis Ababa, Ethiopia
4. Department of Medical Biology, Unit of Microbiology, University of Kinshasa, Kinshasa, Democratic Republic of the Congo
# Contributed equally to this work
Data availability
Genome sequences are available here
Acknowledgment
The regional sequencing and training was coordinated and supported by the Africa CDC Africa Pathogen Genomics Initiative (Africa PGI). Africa PGI acknowledges the support of the African Union, BMGF, FIND, and CDC for this project.
Disclosure and contact details:
The genomes of DENV from Chad are being shared pre-publication to support the public health response. Data may be used and analyzed for these purposes. A scientific publication is in preparation. If you intend to use these sequences prior to our publication, please contact Dr. Djamal Hachim-Abdoulaye ([email protected]) and/or Prof. Placide Mbala-Kingebeni ([email protected]).
References
-
World Health Organization. Disease Outbreak News: Dengue-Chad. 2023 [cited 2023 December 12th]. Available from: Dengue - Chad.
-
Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12(6):1261-76. Epub 20170524. doi: 10.1038/nprot.2017.066. PubMed PMID: 28538739; PubMed Central PMCID: PMCPMC5902022.
-
Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022-7. doi: 10.1093/molbev/msab120. PubMed PMID: 33892491; PubMed Central PMCID: PMCPMC8233496.