Dear all,
I am sharing the linked statement on the name of Prof. Steve Ahuka-Mundeke, from INRB on the characterization of the ebolavirus genomes causing the outbreaks in the Democratic Republic of Congo.
Thanks,
-Gustavo Palacios
Director, Center for Genome Sciences, USAMRIID
-Prof. Steve Ahuka-Mundeke ([email protected])
INRB, School of Medicine Kinshasa University
https://drive.google.com/open?id=1Ih02T5z1cpqEgsBUsLtOIQhuEeb2Yh5Y
INRB Statement
From May 8th to July 25th, 2018, the Democratic Republic of the Congo faced its ninth Ebola
outbreak, which affected 54 people, including 33 deaths in the health zones of Bikoro, Iboko and
Wangata, in the Province of Equateur. Institut National de Recherche Biomedicale (INRB), in
collaboration with its partners, deployed 3 mobile laboratories on the 3 affected sites (Mbandaka,
Bikoro and Itipo) to contribute quickly and efficiently to control this epidemic. A total of 432 samples were collected from these 3 laboratories, of which 51 were positive in real-time RT-PCR. These came from the 38 confirmed cases for the whole period of the epidemic (8 May- 25 July 2018). On
July 10th, samples from the mobile laboratories arrived at INRB, and processing of samples and
sequencing began. Thus far 23 samples have been processed yielding 15 coding-complete
genomes that will be released shortly on GenBank as well. Initial phylogenetic analysis indicates
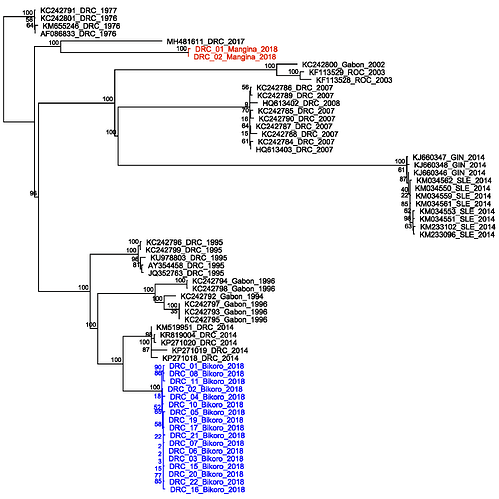
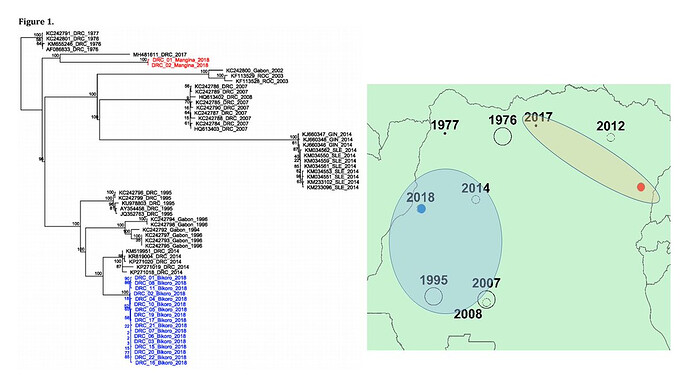
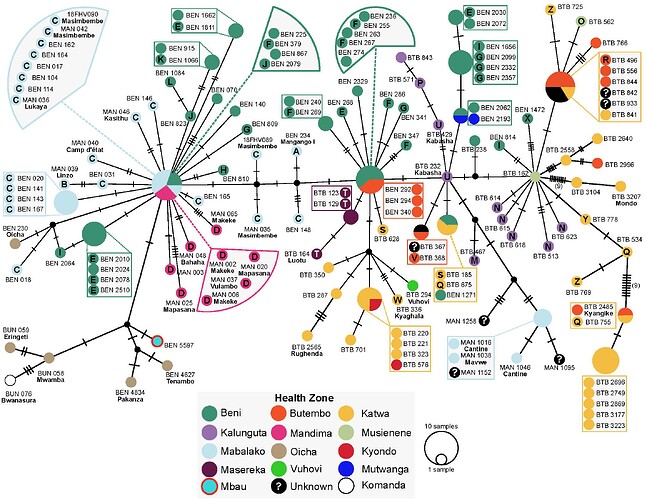
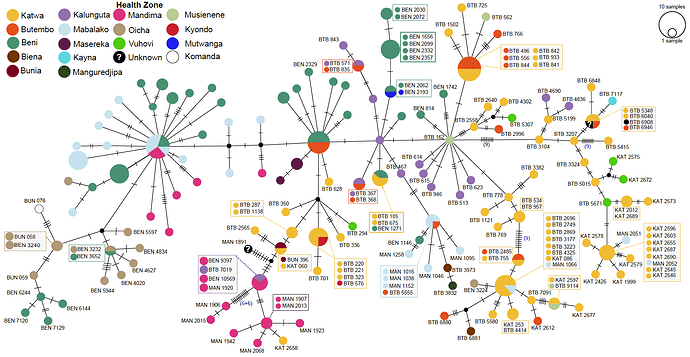
these genomes are closest to the 2014 DRC outbreak (Figure 1, blue labels). Further analysis will
need to be conducted on these preliminary findings as the main focus is to finish processing
available samples from the outbreak, which will be made available as soon as they are generated.
Unfortunately, on August 1st a new cluster of cases was discovered in the region of North
Kivu and confirmed at INRB. Samples were prepared for sequencing at INRB. Two full genome
sequences were obtained (Figure 1, red labels).
Figure 1
The data confirms that the current EVD cases in Nord Kivu (July 30th and ongoing), DRC are caused by a distinct EBOV “strain” than the one causing the outbreak in Equateur province. Based on this findings, although both events are caused by the same ebolavirus species (EBOV), the outbreaks appear not to be connected. On that basis, the available vaccine should work against the new strain.
Partners and collaborators
This work is part of ongoing research collaborations between several partners:
Institut National de Recherche Biomédicale (INRB)
US Army Medical Research Institute of Infectious Diseases (USAMRIID)
Institut Pasteur de Dakar, Dakar, Senegal (IPD)
TransVIHMI (IRD, INSERM, University of Montpellier)
Data availability
Genome sequences for all samples had been shared with all local and international partners working in the DRC. The information reported here is being shared pre-publication to help the research and public health communities respond to the current Ebola virus disease outbreak. Please note though that this data is still based on work in progress and should be considered preliminary. Our analysis of this data is ongoing and a publication communicating our findings is in preparation. INRB will distribute these data to any entity or collaborator that needs it. Please communicate with Drs. Muyembe-Tamfum and Ahuka for further coordination.
Prof. Jean-Jacques Muyembe Tamfum (INRB, School of Medicine Kinshasa University)
Prof. Steve Ahuka-Mundeke (INRB, School of Medicine Kinshasa University)
Dr. Gustavo Palacios (USAMRIID)
Dr. Michael Wiley (USAMRIID)
Dr. Amadou Sall (Institut Pasteur de Dakar, Dakar, Senegal)
Dr. Ousmane Faye (Institut Pasteur de Dakar, Dakar, Senegal)
Prof. Eric Delaporte (TransVIHMI (IRD, INSERM, University of Montpellier)
Dr. Martine Peeters (TransVIHMI (IRD, INSERM, University of Montpellier)
Avenue de la Démocratie N°5345 – Kin/Gombe – B.P. 1197 – KINSHASA 1 – Tél. 898949 289 – 9913614 - 99 23123 – 081 5144285 E-mail : [email protected], Site web : www.inrb.cd