Estimates of the clock and TMRCA for 2019-nCoV based on 27 genomes
January 25, 2020
Kristian Andersen, Scripps Research
[email protected]
Following up on the analyses provided by Andrew Rambaut this is a brief report estimating the evolutionary rate and timing of the epidemic (date of the most recent ancestor (MRCA)) based on 27 publicly shared n2019-nCoV genome sequences. Compared to earlier analyses where several parameters had to be fixed, there is now enough information content in the sequences to obtain reasonable estimates of the clock and TMRCA without fixing parameters. This work is for information purposes only and is not intended for publication. All the data used here is provided by the laboratories listed below through NCBI Genbank or GISAID.
Data
As of January 25, 2020, 28 full-length nCoV-2019 genomes and 1 partial genome are available on the GISAID platform. The partial genome (EPI_ISL_402126) and one with too many sequencing errors (EPI_ISL_403928) were eliminated from these analyses. The final dataset contained 27 full-length nCoV-2019 genomes with 41 SNPs in total, 9 of them masked because of likely sequencing errors (leaving 32 SNPs in the dataset). Acknowledgements of the genome sequences used in this analysis are in the table at the end of this document.
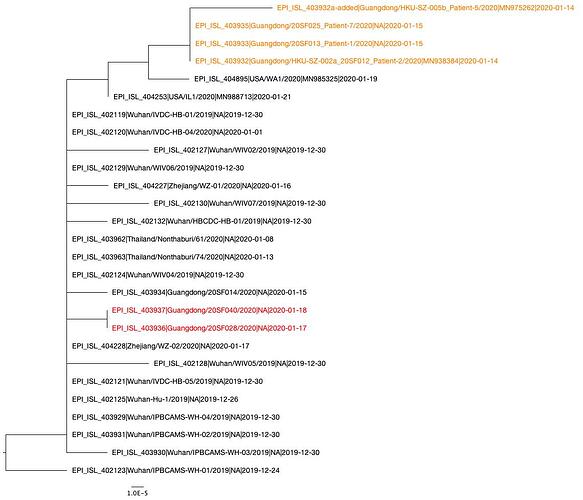
Phylogenetic Tree
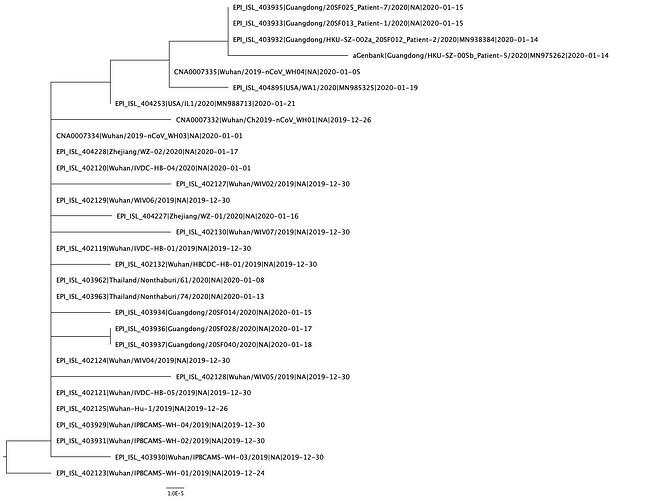
A phylogenetic tree was created using PhyML and in agreement with previous analyses, still shows limited genetic variation in the sampled viruses, which is consistent with a recent common ancestor. Two distinct clusters can also be seen from the tree, consistent with reported human clusters of cases (shown in orange and red).
We are starting to see more structure in the tree and overall the genetic data is highly suggestive of a single-point introduction into the human population followed by sustained human-to-human transmission. This introduction was likely via either a single infected animal or a small cluster of recently infected animals directly into either a single human individual or a small cluster of human individuals. All subsequent cases are the result of human-to-human transmission with no further evidence of zoonotic transmissions.
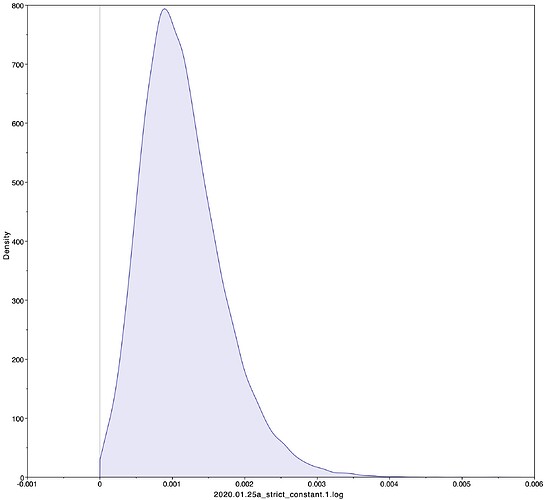
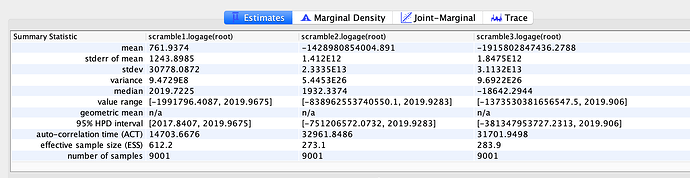
Evolutionary rate
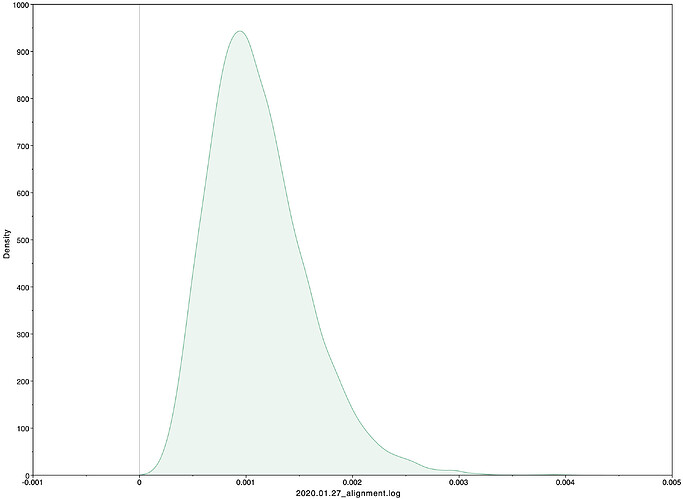
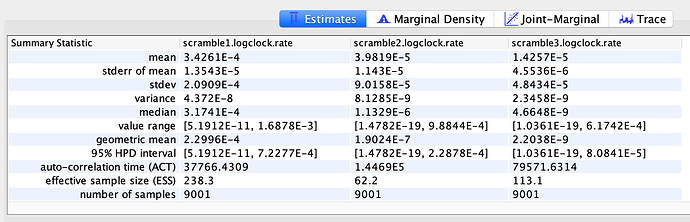
To estimate the substitution rate of nCoV-2019, I used BEAST with a simple model consisting of HKYγ, strict clock with a CTMC rate prior, and a constant tree prior. The median estimate for the substitution rate is very similar to other RNA viruses, including SARS-CoV, Ebola virus, Zika virus, and others at ~ 1E-3 subs/site/year. The range is still wide, but should improve as more sequence data is produced.
|Median|95% HPD
|—|—|—|—|
1.067E-3 | 4.031E-6 - 5.53E-3
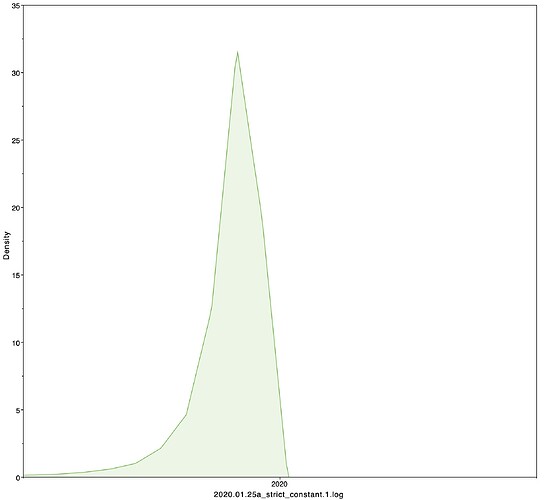
Date of the MRCA
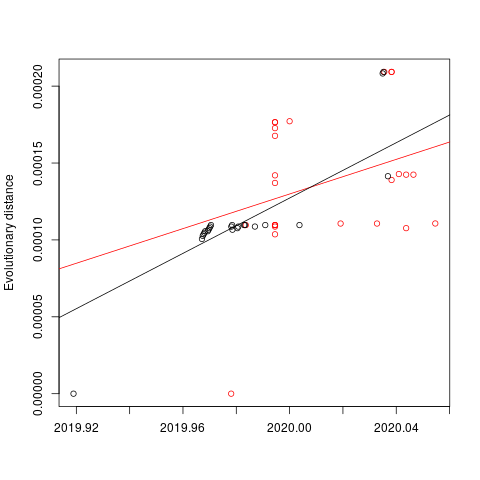
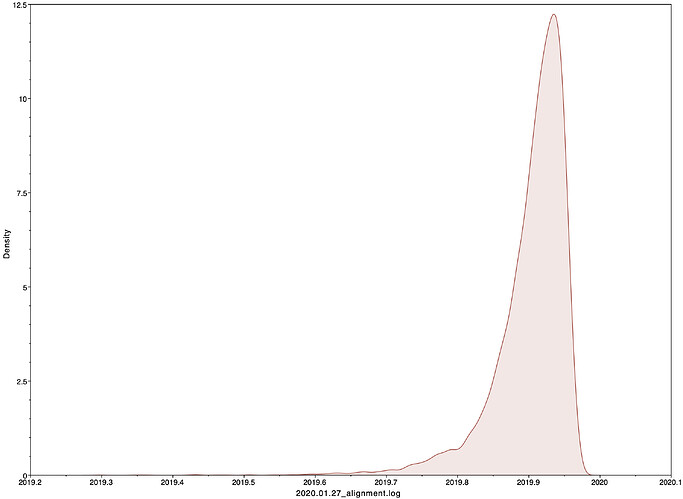
I next estimated the date of the MRCA of the sampled nCoV-2019 genomes, corresponding to the point of the ancestral virus of all the sampled cases was in the same host - in other words, the initial spillover event leading to the outbreak. The first case symptoms was recently reported to be December 1, 2019, although WHO has previously reported this as December 8, 2019. The estimate from BEAST is in agreement with these dates, giving a median date of December 2, 2019. This date is also consistent with prior phylogenetic analyses using fixed rates of the evolutionary rate of 2019-nCoV.
|Median|95% HPD
|—|—|—|—|
02 Dec 2019 | 01 Oct 2019 - 22 Dec 2019
Caveats
Earlier versions of our alignments had significant issues with sequencing errors (I estimate up to 50%). I believe that this issue is minimized in this dataset, with only 9/41 SNPs looking suspicious (and therefore masked in these analyses). That said, there is still limited variation in the sampled genomes and even small artefacts and sequencing errors could greatly influence the estimates.
The clock and TMRCA estimates have large intervals and the median values should be interpreted with caution. The ranges are more appropriate for interpretation of the dates, as opposed to any one of point media values mentioned above. They are all likely to change - possibly significantly - as more patients are sampled and genomes produced.
Acknowledgements and Genome Availability
| Strain | Authors | Source | Lab |
|---|---|---|---|
| EPI_ISL_402119 | Wenjie Tan, et al. | GISAID | National Institute for Viral Disease Control and Prevention, China CDC |
| EPI_ISL_402120 | Wenjie Tan, et al. | GISAID | National Institute for Viral Disease Control and Prevention, China CDC |
| EPI_ISL_402121 | Wenjie Tan, et al. | GISAID | National Institute for Viral Disease Control and Prevention, China CDC |
| EPI_ISL_402123 | Lili Ren, et al. | GISAID | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| EPI_ISL_402124 | Peng Zhou, et al. | GISAID | Wuhan Institute of Virology, Chinese Academy of Sciences |
| EPI_ISL_402125 | Zhang, et al. | GISAID | National Institute for Communicable Disease Control and Prevention (ICDC) Chinese Center for Disease Control and Prevention (China CDC) |
| EPI_ISL_402127 | Peng Zhou, et al. | GISAID | Wuhan Institute of Virology, Chinese Academy of Sciences |
| EPI_ISL_402128 | Peng Zhou, et al. | GISAID | Wuhan Institute of Virology, Chinese Academy of Sciences |
| EPI_ISL_402129 | Peng Zhou, et al. | GISAID | Wuhan Institute of Virology, Chinese Academy of Sciences |
| EPI_ISL_402130 | Peng Zhou, et al. | GISAID | Wuhan Institute of Virology, Chinese Academy of Sciences |
| EPI_ISL_402132 | Bin Fang, et al. | GISAID | Hubei Provincial Center for Disease Control and Prevention |
| EPI_ISL_403929 | Lili Ren, et al. | GISAID | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| EPI_ISL_403930 | Lili Ren, et al. | GISAID | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| EPI_ISL_403931 | Lili Ren, et al. | GISAID | Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College |
| EPI_ISL_403932 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403933 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403934 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403935 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403936 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403937 | Min Kang, et al. | GISAID | Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention |
| EPI_ISL_403962 | Pilailuk, et al. | GISAID | Department of Medical Sciences, Ministry of Public Health, Thailand |
| EPI_ISL_403963 | Pilailuk, et al. | GISAID | Department of Medical Sciences, Ministry of Public Health, Thailand |
| EPI_ISL_404227 | Yin Chen, et al. | GISAID | Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention |
| EPI_ISL_404228 | Yanjun Zhang, et al. | GISAID | Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention |
| EPI_ISL_404253 | Ying Tao, et al. | GISAID | IL Department of Public Health Chicago Laboratory |
| EPI_ISL_404895 | Queen, et al. | GISAID | Division of Viral Diseases, Centers for Disease Control and Prevention |
| MN975262 | Chan et al. | Genbank | State Key Laboratory of Emerging Infectious Diseases |