Liu et al. showed that A to V amino acid substitution at position 188 in NS1 enhanced zika virus (ZIKV) infectivity (Liu et al., 2017). It was expected that A to V aa substitution affect the NS1 dimerization. However, the A to V substitution at position 188 does not influcnce NS1 dimerization. Thus the mechanism by which the A to V substitution affects NS1 phenotype remains unclear.
Many RNA viruses including flaviviruses form functional RNA secondary structure in their genomes. Thus we investigated whether the RNA secondary structure was involved in the phenotypic change of NS1 by A to V change.
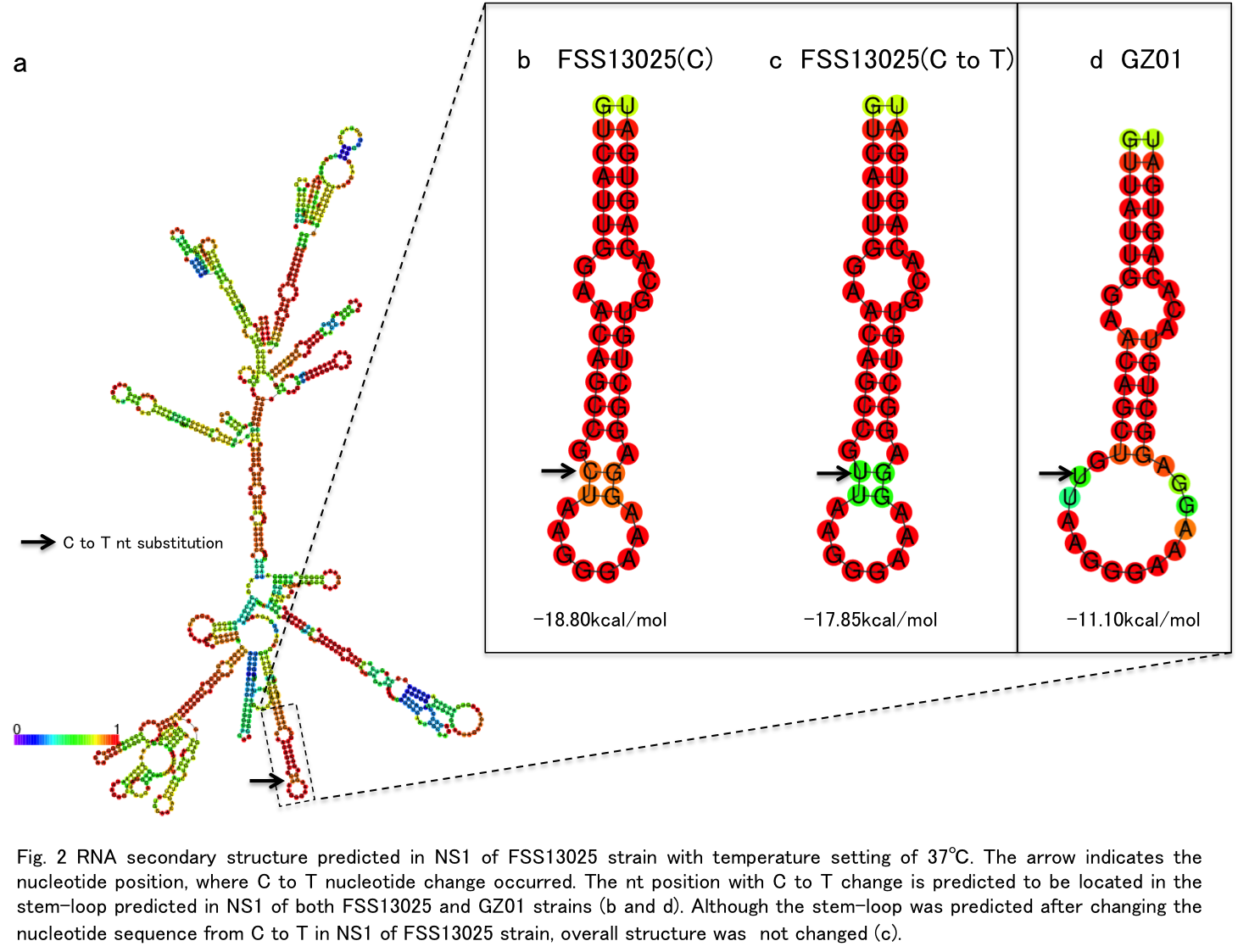
A to V substitution at position 188 is caused by a single nucleotide change from C to T (Fig. 1). When the RNA secondary structure in NS1 of FSS13025 strain was predicted using RNAfold program with temperature setting 37℃, the 188 position was located in the part of stem-loop structure predicted in NS1 (Fig.2a).
However, when the stem-loop was predicted after changing the nucltotide sequence from C to T, overall structure of the stem-loop was not changed (Fig.2 c). This result was not changed even the secondary structure was predicted under the temperaure setting 18℃, which is incubation temperature for mosquito cell line.
Thus the stem-loop predicted here is unlikely to be involvement in the phenotypic change of ZIKV NS1 by C to T change.
Reference
Liu Y, Liu J, Du S, Shan C, Nie K, Zhang R, Li XF, Zhang R, Wang T, Qin CF, Wang P, Shi PY, Cheng G. (2017) Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature. 545:482-486. doi: 10.1038/nature22365.