Nigeria is experiencing a high number of Lassa fever cases this year. A collaboration between the Irrua specialist Teaching Hospital (ISTH), Institute for Lassa Fever Research and Control (ILFRC), Irrua, Edo State, Nigeria, The Bernhard-Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany, Public Health England (PHE), together with additional collaborators and funders, has started to sequence Lassa fever virus (LASV) samples from 2018. Sequencing (MinION technology, Oxford Nanopore), analyses and production of consensus Lassa viruses are being performed in the field at ISTH, ILFRC laboratory. The first 7 consensus sequences for both the L and S segments can be downloaded here:
https://github.com/ISTH-BNITM-PHE/LASVsequencing
The sequences are derived from blood of 7 laboratory-confirmed Lassa fever patients from Edo, Ondo, Ebonyi and Imo States. In addition, unpublished Lassa virus sequences (partial and complete) from previous years, that are under analysis by BNITM, are also made available to facilitate the interpretation of the 2018 sequences. Please note that this is an early release, so accuracy cannot be guaranteed at this stage (see also Disclaimer below).
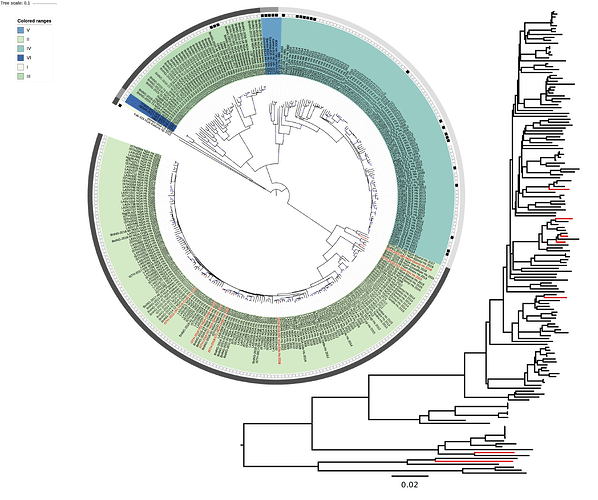
Maximum likelihood phylogenetic analyses of the coding genes in both the L and S segments demonstrate that the 7 2018 viruses (with red label) fall within the Nigerian LASV diversity, specifically lineage II. The 2018 viruses are not directly linked, a pattern that is consistent with spill-over from the rodent reservoir rather than sustained human-to-human transmission.
Figure 1: Lassa virus S-segment phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different color ranges. The rectangular tree is the lineage II subtree (the largest light-green color range in the circular tree). The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. The outer circle represents geographic origin: dark grey = Nigeria, light grey: Mano River Union countries, intermediate grey = other (Togo, Benin & Mali). Newly sequenced viruses are depicted in red. 100% bootstrap support is indicated with a circle at the internal nodes.
Figure 2: Lassa virus L-segment phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different color ranges. The rectangular tree is the lineage II subtree (the largest light-green color range in the circular tree). The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. The outer circle represents geographic origin: dark grey = Nigeria, light grey: Mano River Union countries, intermediate grey = other (Togo, Benin & Mali). Newly sequenced viruses are depicted in red. 100% bootstrap support is indicated with a circle at the internal nodes.
We will try to regularly update both the sequence release and the analyses.
DISCLAIMER
The Irrua Specialist Teaching Hospital (ISTH), Edo State, Nigeria, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany, and Public Health England (PHE), UK, are committed to sharing data in public health emergencies. We release unpublished Lassa virus sequences from Nigeria to support the public health response as well as the development and evaluation of Lassa fever diagnostics or therapeutics. The data may be used and analyzed for these purposes. It is not permitted to use the sequences for publication, i.e. any type of communication with the general public that describes data generated with the use of the sequences. If you intend to so, please contact us directly.
Dr. Ephraim Epogbaini [email protected], Director of Institute of Lassa Fever Research and Control,
Prof. Stephan Günther [email protected], Director, WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Reference and Research, BNITM